Running a multi-stage workflow#
A workflow is a definition of execution of functions. It defines the order of execution of multiple dependent steps in a directed acyclic graph (DAG). A workflow can reference the project’s params, secrets, artifacts, etc. It can also use a function execution output as a function execution input (which, of course, defines the order of execution).
MLRun supports running workflows on a local or kubeflow pipeline engine. The local engine runs the workflow as a
local process, which is simpler for debugging and running simple/sequential tasks. The kubeflow ("kfp") engine runs as a task over the
cluster and supports more advanced operations (conditions, branches, etc.). You can select the engine at runtime. Kubeflow-specific
directives like conditions and branches are not supported by the local engine.
Workflows are saved/registered in the project using the set_workflow().
Workflows are executed using the run() method or using the CLI command mlrun project.
Refer to the Tutorials and Examples for complete examples.
In this section
Composing workflows#
Workflows are written as python functions that make use of function operations (run, build, deploy)
and can access project parameters, secrets, and artifacts using get_param(), get_secret() and get_artifact_uri().
For workflows to work in Kubeflow you need to add a decorator (@dsl.pipeline(..)) as shown below.
Example workflow:
from kfp import dsl
import mlrun
from mlrun.model import HyperParamOptions
funcs = {}
DATASET = "iris_dataset"
in_kfp = True
@dsl.pipeline(name="Demo training pipeline", description="Shows how to use mlrun.")
def newpipe():
project = mlrun.get_current_project()
# build our ingestion function (container image)
builder = mlrun.build_function("gen-iris")
# run the ingestion function with the new image and params
ingest = mlrun.run_function(
"gen-iris",
name="get-data",
params={"format": "pq"},
outputs=[DATASET],
).after(builder)
# train with hyper-parameters
train = mlrun.run_function(
"train",
name="train",
params={"sample": -1, "label_column": project.get_param("label", "label"), "test_size": 0.10},
hyperparams={
"model_pkg_class": [
"sklearn.ensemble.RandomForestClassifier",
"sklearn.linear_model.LogisticRegression",
"sklearn.ensemble.AdaBoostClassifier",
]
},
hyper_param_options=HyperParamOptions(selector="max.accuracy"),
inputs={"dataset": ingest.outputs[DATASET]},
outputs=["model", "test_set"],
)
print(train.outputs)
# test and visualize our model
mlrun.run_function(
"test",
name="test",
params={"label_column": project.get_param("label", "label")},
inputs={
"models_path": train.outputs["model"],
"test_set": train.outputs["test_set"],
},
)
# deploy our model as a serverless function, we can pass a list of models to serve
serving = mlrun.import_function("hub://v2_model_server", new_name="serving")
deploy = mlrun.deploy_function(
serving,
models=[{"key": f"{DATASET}:v1", "model_path": train.outputs["model"]}],
)
# test out new model server (via REST API calls), use imported function
tester = mlrun.import_function("hub://v2_model_tester", new_name="live_tester")
mlrun.run_function(
tester,
name="model-tester",
params={"addr": deploy.outputs["endpoint"], "model": f"{DATASET}:v1"},
inputs={"table": train.outputs["test_set"]},
)
Note
For defining the steps order you can either use steps outputs as written above, or use .after(step_1,step_2,..) method, that allows the user to define the order of the workflow steps without the need to forward the outputs from the previous steps.
Saving workflows#
If you want to use workflows as part of an automated flow, save them and register them in the project.
Use the set_workflow() method to register workflows, to specify a workflow name,
the path to the workflow file, and the function handler name (or it looks for a handler named "pipeline"), and can
set the default engine (local or kfp).
When setting the embed flag to True, the workflow code is embedded in the project file (can be used if you want to
describe the entire project using a single YAML file).
You can define the schema for workflow arguments (data type, default, doc, etc.) by setting the args_schema with a list
of EntrypointParam objects.
Example:
# define agrument for the workflow
arg = mlrun.model.EntrypointParam(
"model_pkg_class",
type="str",
default="sklearn.linear_model.LogisticRegression",
doc="model package/algorithm",
)
# register the workflow in the project and save the project
project.set_workflow("main", "./myflow.py", handler="newpipe", args_schema=[arg])
project.save()
# run the workflow
project.run("main", arguments={"model_pkg_class": "sklearn.ensemble.RandomForestClassifier"})
Running workflows#
Use the run() method to execute workflows. Specify the workflow using its name
or workflow_path (path to the workflow file) or workflow_handler (the workflow function handler).
You can specify the input arguments for the workflow and can override the system default artifact_path.
Workflows are asynchronous by default. You can set the watch flag to True and the run operation blocks until
completion and prints out the workflow progress. Alternatively, you can use .wait_for_completion() on the run object.
The default workflow engine is kfp. You can override it by specifying the engine in the run() or set_workflow() methods.
Using the local engine executes the workflow state machine locally (its functions still run as cluster jobs).
If you set the local flag to True, the workflow uses the local engine AND the functions run as local process.
This mode is used for local debugging of workflows. The remote engine runs the workflow from a remote pod. From the project
source you can set the remote engine to run in local by setting engine to remote:local.
When running workflows from a git enabled context it first verifies that there are no uncommitted git changes
(to guarantee that workflows that load from git do not use old code versions). You can suppress that check by setting the dirty flag to True.
Examples:
# simple run of workflow 'main' with arguments, block until it completes (watch=True)
run = project.run("main", arguments={"param1": 6}, watch=True)
# run workflow specified with a function handler (my_pipe)
run = project.run(workflow_handler=my_pipe)
# wait for pipeline completion
run.wait_for_completion()
# run workflow in local debug mode
run = project.run(workflow_handler=my_pipe, local=True, arguments={"param1": 6})
Notification#
Instead of waiting for completion, you can set up a notification in Slack with a results summary, similar to:
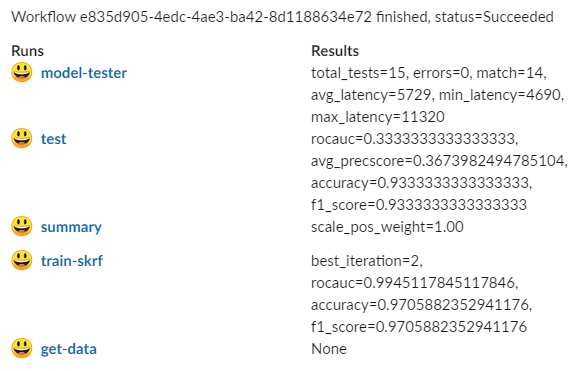
Use one of:
project.notifiers.add_notification(notification_type="slack",params={"webhook":"<user-slack-webhook>"})
or in a Jupyter notebook with the %env magic command:
%env SLACK_WEBHOOK=<slack webhook url>
